Pisum Genetics
2008—Volume 40
Research Papers
Mapping of Sym28
Significant linkage was found between Sym28 and LGV markers. Based on the linkage data, a map fragment surrounding Sym28 was constructed (Fig. 1b). The arrangement of sequence-tagged markers (CAPS) in the resulting map was compared with the positions of putative homologous loci (according to BLAST search results) on the physical map of Medicago truncatula Gaertn. chromosome 7 (Fig. 1c) which was reported to be syntenic with LGV of pea (4). The obtained map was also compared with the one containing Nod4 and morphological markers, as reported in (11). Map position of genes Sym28 and Nod4 was similar. Obviously, more linkage analyses are needed to reveal a more certain position of Nod4 in relation with Sym28 and possible correspondence between the genes studied in this report and known genes controlling meristem identity in Arabidopsis.
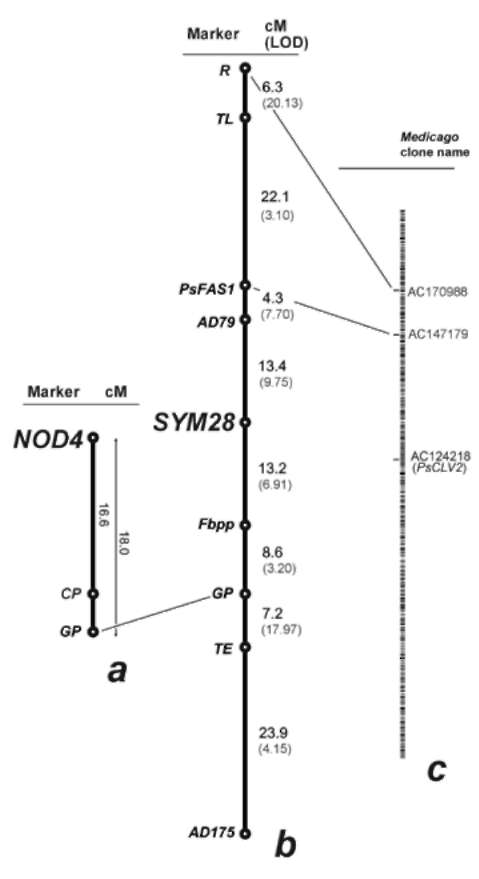
Figure 1. Correspondence between regions of pea LGV containing genes nod4 (a, after (11)), sym28 (b) and a physical map of chromosome 7 of Medicago truncatula (c) generated by Medicago CviT- BLAST tool (http://www.medicago.org/genome/cvit_blast.php). Chromosome orientation was established according to (13). Numbers between markers designate distances, cM; numbers in parentheses stand for LOD score.
Acknowledgements: The authors are indebted to Dr. Viktor E. Tsyganov (All-Russia Research Institute of Agricultural Microbiology) and to Dr. Klavdia K. Sidorova (Institute for Cytology and Genetics ňî÷íĺĺ) for generously provided seeds of studied lines P64 and K301, respectively. The authors also thank Olga A. Ash and Galina A. Khartina for assistance in field work. The work was supported by Russian Foundation for Basic Research (grant no. 07-04-00652).
17