Pi sum Genetics
2003—Volume 35
Research Papers
Genome walking in pea:
an approach to clone unknown flanking sequences
an approach to clone unknown flanking sequences
Chawla, R. and
DeMason, D.
DeMason, D.
Botany and Plant Sci.
Univ. of California, Riverside, CA 92507, USA
Univ. of California, Riverside, CA 92507, USA
The isolation and characterization of unknown DNA sequences flanking known regions are critical,
especially for the analysis of upstream and downstream noncoding regions. The traditional approach for
'walking' from regions of known sequence into flanking DNA sequences involves the successive probing of
libraries with clones obtained from prior screenings. This method of screening DNA libraries is a relatively
time consuming procedure and requires the use of radioactive probes. Advancements in the PCR technique
have helped researchers to reduce this time and avoid the use of radioactive probes (1). Genome walking is a
relatively fast, reliable and general approach to sequence or clone DNA adjacent to a known region.
especially for the analysis of upstream and downstream noncoding regions. The traditional approach for
'walking' from regions of known sequence into flanking DNA sequences involves the successive probing of
libraries with clones obtained from prior screenings. This method of screening DNA libraries is a relatively
time consuming procedure and requires the use of radioactive probes. Advancements in the PCR technique
have helped researchers to reduce this time and avoid the use of radioactive probes (1). Genome walking is a
relatively fast, reliable and general approach to sequence or clone DNA adjacent to a known region.
Promoters are segments of DNA that regulate the timing and location of gene expression. The promoter
sequence is usually located upstream of the transcription start site, but regulatory elements can be present in
5' untranslated regions (UTRs), within introns or in the 3' UTRs of genes. Analysis of promoter sequences in
combination with new databases like PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/fasta.html) and
PlantCARE (http://oberon.fvms.ugent.be:8080/PlantCARE/index.html) provide possible insights into the
regulation of important plant genes. One very powerful way of modifying the characteristics of plants is to
target the expression of introduced genes to specific parts of the plant or at specific stages of the life cycle
using promoters with known specificity. Very few pea promoters are currently known. The goal of this paper
is to summarize our success with cloning promoter sequences of pea genes using the technique of genome
walking. We discuss the method by
which it works and the results obtained.
sequence is usually located upstream of the transcription start site, but regulatory elements can be present in
5' untranslated regions (UTRs), within introns or in the 3' UTRs of genes. Analysis of promoter sequences in
combination with new databases like PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/fasta.html) and
PlantCARE (http://oberon.fvms.ugent.be:8080/PlantCARE/index.html) provide possible insights into the
regulation of important plant genes. One very powerful way of modifying the characteristics of plants is to
target the expression of introduced genes to specific parts of the plant or at specific stages of the life cycle
using promoters with known specificity. Very few pea promoters are currently known. The goal of this paper
is to summarize our success with cloning promoter sequences of pea genes using the technique of genome
walking. We discuss the method by
which it works and the results obtained.
Materials and Methods
Plant materials
The pea genotype used in this study
is from the Marx collection, which
resides in the USDA Western Regional
Plant Introduction Station. It is W6
22593, which is designated as WT or Af
St Tl. Seeds were sown in UC soil mix
supplemented with slow release fertilizer
in 1 gallon pots and plants were grown
under standard greenhouse conditions
and natural light regimes. Leaves of 1-
month old plants were frozen at -80 C
until DNA extraction.
is from the Marx collection, which
resides in the USDA Western Regional
Plant Introduction Station. It is W6
22593, which is designated as WT or Af
St Tl. Seeds were sown in UC soil mix
supplemented with slow release fertilizer
in 1 gallon pots and plants were grown
under standard greenhouse conditions
and natural light regimes. Leaves of 1-
month old plants were frozen at -80 C
until DNA extraction.
The GenomeWalking technique
The GenomeWalking technique is
summarized in Fig. 1. First, genomic
DNA is isolated from plant tissues. The
quality of DNA is checked for high
average molecular weight on a 0.8%
agarose gel. The starting DNA must be
very clean and have a high average
summarized in Fig. 1. First, genomic
DNA is isolated from plant tissues. The
quality of DNA is checked for high
average molecular weight on a 0.8%
agarose gel. The starting DNA must be
very clean and have a high average
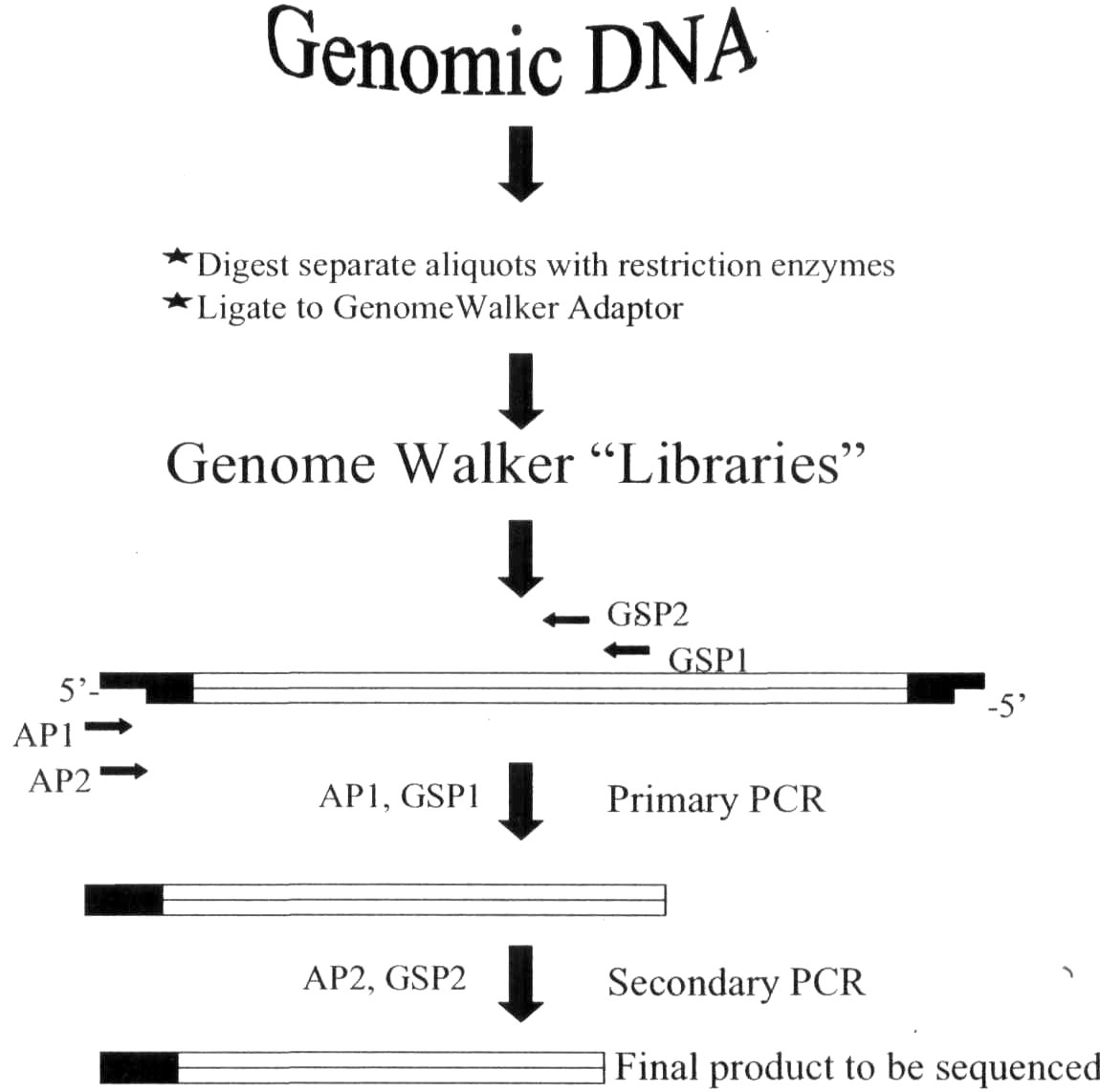
Fig. 1. Flow chart of the GenomeWalker protocol. AP1 and AP2
represent adaptor primers and GSP1 and GSP2 represent gene
specific primers.
represent adaptor primers and GSP1 and GSP2 represent gene
specific primers.