
|
Pisum Genetics_____________________________2003—Volume 35_______________________Research
Papers
The Pisum genus has only one recessive gene for powdery mildew resistance
Sharma, B. Div. of Gen., Indian Agric. Res. Inst.
Delhi, India
Powdery mildew resistance (PMR) in pea displays several interesting aspects. In a great majority of plant species, powdery mildew resistance is controlled by a dominant allele and, therefore, stands a chance of breakdown through mutation of the gene(s) into recessive state. In pea the resistance is produced by a recessive allele, and reversion of recessive alleles to dominant alleles is not known in plants. Moreover, point mutations revert very rarely even in microbial systems, and recessive mutations caused by deletions have no possibility to revert. The recessive gene for PMR in pea is very stable in its action over time and space and imparts resistance against all forms of the Erysiphe pisi fungus. Most importantly, there appears to be only one locus for PMR in the various taxa within the genus Pisum, which is the subject of the present discussion. The situation is qualitatively different from other plants where this trait is under multigenic control.
Although the first publication on the genetics of PMR of pea appeared in 1925 (2), it was Harland (3) who convincingly demonstrated the monogenic recessive nature of the trait two decades later. Heringa et al. (4), on the other hand, concluded PMR to be under the control of two unlinked recessive genes. Several other studies (e.g. 7, 15, 16) reported multigenic control of this trait. However, such conclusions were generally not convincing, sometimes self-contradictory (7, 15), sometimes based on vague generalizations. Based on the two-gene theory, the genes proposed were erl (the original Harland's) and er2. Over a period of 25 years of pea breeding for PMR, we never observed anything other than the monogenic nature of this character. What could be the reason? The situation can be reviewed on the basis of experience and the information generated from Ph. D. programs (1, 5, 6, 10, 11) and other studies (12, 13, 14, and unpublished).
It was found that PMR has both quantitative and qualitative expression simultaneously, which causes erroneous scoring in a segregating population, leading to variable F2 ratios. It was noticed long ago that the recessive gene allows a certain degree of fungal growth on the PMR genotypes, which however is restricted to the foliage, i.e. leaves and stipules. In the Indian plains during winter, such infection is very sporadic, highly unpredictable, and extremely variable depending on climatic conditions, genotypic constitution, and age of plants. The fungus becomes apparent only toward the end of plant life. High humidity in combination with warm days and cooler nights favors fungal growth, early varieties catch infection earlier, and old plants succumb to the onslaught of the fungus while young plants of the same genotype remain infection-free even up to flowering stage in the same conditions (observations in summer crop at Lahaul). The fungal growth on the foliage of PMR genotypes has quantitative expression at different locations, in different crop seasons, and on different days of the same season as the crop advances.
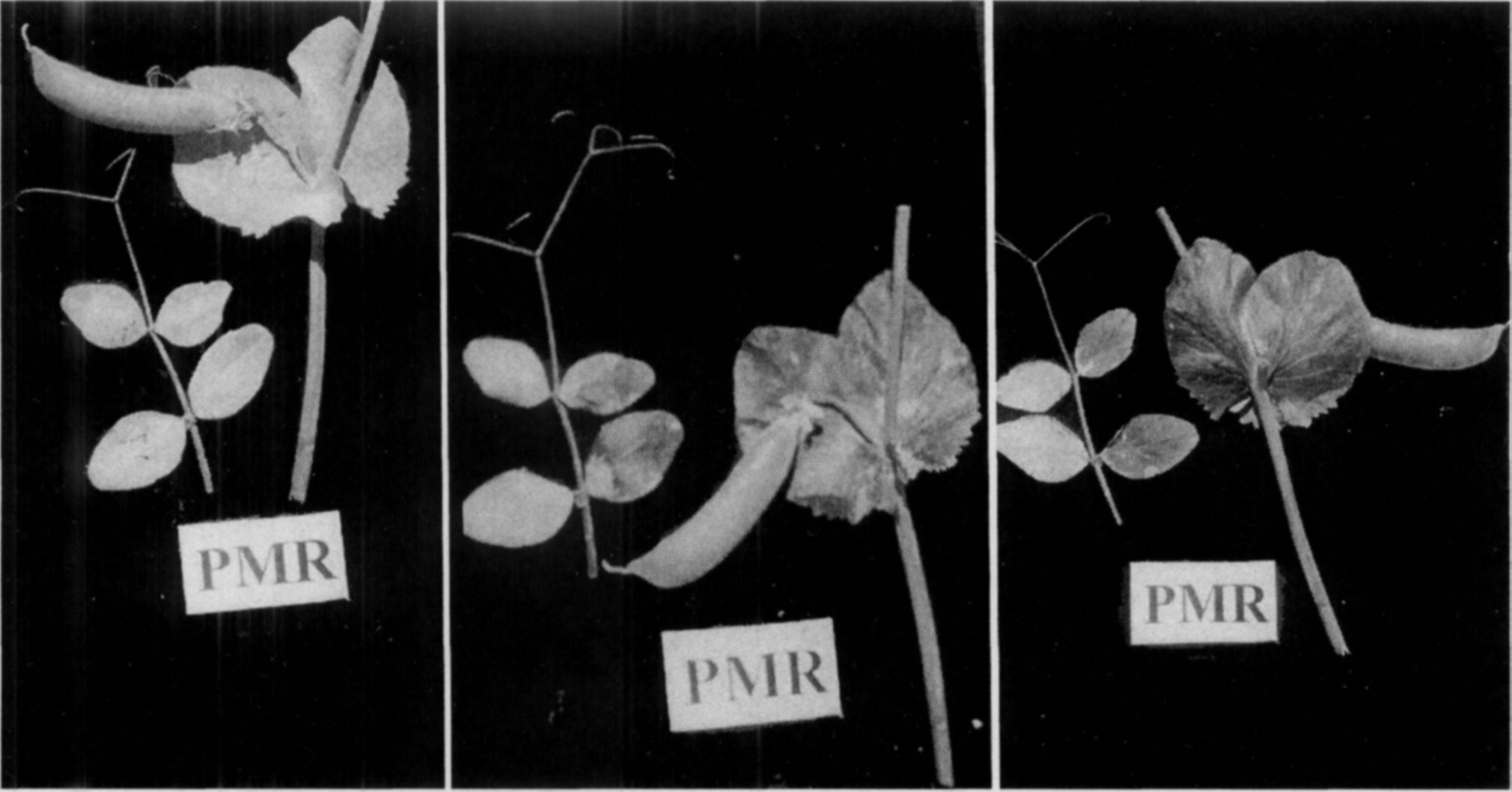
The qualitative aspect of the infection is that whatever the degree of fungal growth on the foliage of PMR plants, the stem, peduncles, and pods are totally free of infection (Fig.l). Another qualitative difference is that the epidermal tissue even in the fungus covered areas of leaves in the PMR plants remains healthy under the mycelial cover and does not turn brown, whereas the infected surface of PMS genotypes becomes black by the time the plants mature. The leaf surface exposed after wiping the fungal coat from the leaves of PMR plants is normal and healthy as if it had no infection (Fig. 2). The distinction between PMS and infected PMR plants is unmistakable even when the crop is dried. Dry plants of PMR genotypes look clear white while those of PMS strains become blackish with sickly appearance. Such infection causes no economic losses in the PMR strains despite severe fungal growth on the foliage of green plants. If fungicidal sprays are used to keep plants disease free, no perceptible change in the productivity of PMR cultivars is observed while the grain yields of PMS strains increases nearly 1.5 fold (12). The grain quality of the susceptible genotypes also improved to normal level.
|
 |
||
|
|
||
|
Fig. 1. Growth of Erysiphe pisi on the plants of powdery mildew susceptible (left) and resistant (right) plants. Intensity of fungal growth on the foliage of both plants being almost similar, the stem, peduncles, and pods of the resistant plant are totally free of infection.
|
||
|
|
||
 |
||
|
|
||
|
Fig. 2. Effect of powdery mildew infection on the leaves of PMR plants. Left—thick and uniform fungal growth covering the foliar surface. Fungus partially wiped off the ventral (middle) and dorsal (right) surfaces of leaves and stipules. Note the healthy tissue exposed despite heavy infection.
|
|
The uncertainties associated with disease development in the plains are eliminated when pea strains are grown during summer in the Lahaul Valley of Himachal Pradesh in high Himalayas (altitude about 4000 m) where the nights are cool, days warm, and rainfall more than during winters in the plains. The location serves as a hotspot for powdery mildew in pea, lentil, wheat and other crops. It occurs universally in high intensity, and the PMR genotypes always develop fungus on the foliage to variable degree. This facility has been utilized to confirm the PMS/PMR status of doubtful genotypes, as there are chances of error in the plains in the crop seasons of low intensity or late development of the disease. The genetic analysis for Er in all our studies was done distinguishing the PMR and PMS plants on the basis of infection on the stem, peduncles and pod, ignoring the fungal growth, whatever its intensity, on the foliage. The results of a series of studies summarized here do not support a digenic model for PMR in pea.
Data of five crosses (1) were analyzed for monogenic, two patterns of digenic, and trigenic segregations (Table 1). Deliberately, the disease scoring was done on a 5-point scale following the procedure usually adopted in field, which is based on gross visual assessment. This approach contains an inherent risk of misclassification between grade 1 (resistant) and grade 2 (susceptible) which, like any neighboring grades, are prone to subjective evaluation of individual plants in a segregating population. The problem will be further compounded if the parents differ in maturity duration and the F2 plants are in a wide range of developmental stages at any point of crop growth. The early PMR segregants with higher fungal intensity on foliage are more likely to be placed in the susceptible range. With such errors, even if rare, the chances of observing different segregation ratios increase. As can be seen from the results presented in Table 1, the F2 data do not fit any of the four ratios of monogenic, digenic, and trigenic sgregation. Any ratio could have been obtained in such a situation.
Table 1. Segregation in the F2 population of five crosses scored for powdery mildew infection on 1-5 point scale (Grades: 1—resistant; 2-5—susceptible with varying degree of disease intensity)
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
With this experience, screening for PMS-PMR reaction was thereafter done on the basis of fungal infection on the stem, peduncle, and pods. Analysis of a pooled population of 41,277 plants (Table 2), which included observations recorded at Delhi as well as Lahaul, resulted in a near-perfect segregation into 3: 1 ratio (31,101 S: 10,176 R).
Table 2. Segregation for powdery mildew resistance in different studies
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
The progenies of eleven test-crosses (Table 3), without exception, segregated in 1 S : 1 R ratio (1), which also confirms the monogenic inheritance of PMR. Nonsignificant heterogeneity x2 allows us to pool the data of the crosses. Segregation of the pooled back-crossed population into 201 susceptible and 190 resistant plants (x2 = 0.31, P = 0.58) provides strong support to this conclusion. All the six PMR strains used in this study were developed at or collected from different centers in India (DMR 11, HUP 5, HFP 4), Canada (Tara), U.K. (S 143), and Mexico (Mexique 4) and do not share pedigree. In all probability, they had different parents as the PMR donor. In the present context, Mexique 4, a strain collected from nature in Mexico, is of greatest relevance. It was used by Heringa (4) to propose two-gene control of PMR. However, pooled analysis of the test-crosses of Mexique 4 with four different PMS cultivars resulted in segregation with a very good fit to 1:1 ratio. The total population of 143 plants segregated into 74 S and 69 R back-crossed plants [x2 (1 : 1) = 0.175]. This result is too far away from the 1 S : 3 R ratio (36 S : 107 R) expected if%two genes with independent assortment were involved in causing powdery mildew resistance in pea. Four crosses of Mexique 4 with the four totally unrelated PMS strains (Pusa 10—developed through hybridization at IARI in the late seventies, Type 163—a local selection from farmers' field, under cultivation in India since the fifties, KPSD 1—Russian variety Flavanda, and PG 3—a cultivar evolved at the Punjab Agricultural University, Gurdaspur, India) segregated into 561 S and 195 R (1). Nonsignificant x2 (0.25 , P = 0.60) for 3 S : 1 R ratio also confirms monogenic nature of PMR trait in Mexique 4. Obviously, any conclusion other than monogenic control of PMR is a result of erroneous classification of the F2 populations. Fungal growth on the foliage of PMR plants ap-pears to be solely responsible for such error. Enough indication to this effect is available even in the present study (Table 1).
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Table 3. Segregation for powdery mildew resistance in test-crosses [(R x S) x R] of pea
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Complementation analysis was also carried out by Gupta (1) using 23 PMR strains of different origin in 45 crosses (Table 4). The results convincingly demonstrate total absence of genetic complementation. The isolated plants marked as PMS do not suggest complementation in F1 or segregation in F2. Such erroneous classification of plants only confirms that even the experienced worker may fail to identify all the plants correctly in a segregating population, especially under heavy fungal growth on the foliage of resistant plants. These observations also confirm that correct conclusions are generally obtained, despite sporadic mistakes, if F2 screening is done with proper understanding about the nature of PMR imparted by the recessive gene er in pea.
Table 4. Complementation for powdery mildew resistance in pea. |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
The hypothesis suggesting a digenic nature to PMR in several reports were never confirmed by a critical test. Marx (8, 9) obtained monogenic segregation for PMR but published conflicting reports about its map position. Initially (8), he placed Er on chromosome 3, and finally (9) concluded the mapping of this gene to be "elusive" while simultaneously questioning the existence of the polymeric gene Er2. It is now clear that the screening procedures,, and not the gene itself, have been the real cause of "elusion". The gene was ultimately mapped on chromosome 6 (11), and tagged with molecular markers (5, 10, 17, 19).
The putative Er2 gene has not been mapped even after twenty four years since its existence was suggested. Tiwari et al. (18) concluded two-gene control of PMR under different growing conditions. However, the authors were not able to locate the second gene, although they successfully tagged Erl on chromosome 6. Very recent studies (13, 14) have revealed that only one gene for powdery mildew resistance appears to be operating in the genus Pisum. These results suggest that there is no justification for maintaining two gene symbols for the PMR trait, and the original symbol Er/er proposed by Harland (3) must be treated as valid. Its recessive nature imparts great stability of expression under diverse conditions over time. It would be interesting to analyze the molecular changes involved in the multiple alleles of different origin. Even more important is to understand how a nonfunctional (or malfunctioning) recessive gene initiates a chain of metabolic reactions that result in a near-perfect protection of the plant from a deadly fungus throughout the world.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
1. Gupta, M.D. 1987. Ph. D. Thesis. Indian Agric. Res. Inst., New Delhi: 160.
2. Hammurlund, C. von. 1925. Hereditas 6: 11-26.
3. Harland, S.C. 1948. Heredity 2: 263-269.
4. Heringa, R.J., Novel, A. van and Tazelaar, M.F. 1969. Euphytica 18: 163-169.
5. Janila, P. 1999. Ph. D. Thesis. Indian Agric. Res. Inst., New Delhi: 95.
6. Kala, Y.K. 1998. Ph. D. Thesis. Indian Agric. Res. Inst., New Delhi: 154.
7. Kumar, H. and Singh, R.B. 1981. Euphytica 30: 147-151.
8. Marx, G.A. 1971. Pisum Newslett. 3: 18-19.
9. Marx, G.A. 1986. Pisum Newslett. 18: 39-41.
10. Rakshit, S. 1997. Ph. D. Thesis. Indian Agric. Res. Inst., New Delhi: 138.
11. Sarala, K. 1993. Ph. D. Thesis. Indian Agric. Res. Inst., New Delhi: 109.
12. Sharma, B. 1995. In.Sharma, B. Kulshreshtha, V.P., Gupta, N. and Mishra, S.K. (eds.) Genetic Research and Education: Current Trends and the Next Fifty Years. Proc. Golden Jubilee Symp. 12-15 February 1991, New Delhi Indian Soc. Genet Plant Breed., New Delhi: 916-924.
13. Sharma, B. 2003. Pisum Genetics, 35: 30-31.
14. Sharma, B. and Yadav, Y. 2003. Pisum Genetics, 35: 31.
15. Singh, R.B., Singh, M.N., Singh U.P. and Singh, R.M. 1983. Indian J. Agric. Sci. 53: 855-859.
16. Sokhi, S.S., Jhooty, J.S. and Bains, S.S. 1979. Indian Phytopath. 32: 571-574.
17. Timmerman, G.M., Frew, T.J., Weeden, N.F., Miller, A.L. and Goulden, D.S. 1994. Theor. Appl. Genet. 88: 1050-1055.
18. Tiwari, K.R., Penner, G.A. and Warkentin, T.D. 1997. Can. J. Plant Sci. 77: 307-310.
19. T iwari, K.R., Penner, G.A. and Warkentin, T.D. 1998. Genome 41: 440-444.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||