Characteristics and inheritance
of the leaf mutation ins
Smirnova, O.G.
Inst. of Cytology and Gen., Russian Acad. of Sci.
Novosibirsk, Russia
Lamprecht was the first to describe insecatus leaves (gene symbol ins)
(1). He studied a pea line with dissected tips of the first pair of leaflets. A
central vein of the incised leaflet is overdeveloped into a small tendril
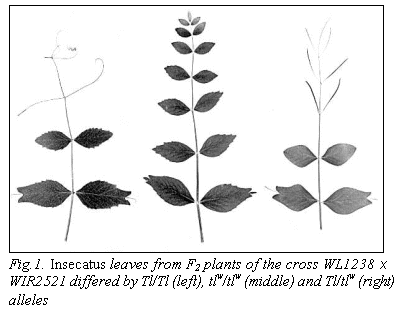
(Fig.1, left).
 In contrast to homeotic pea leaf development mutants afila and tendril-less(tl)
the mutation of gene ins affects only a few compound pea leaves. We have
observed the insecatus phenotype in several lines and determined the late
flowering the insecatus phenotype appearance 2-3 nodes below the first
flowering node. Following initial formation of insecatus leaves, they may
also occur at the next 3-4 nodes. Thus, insecatus leaves are formed in
the middle part of a plant whereas lower and upper nodes of such plants have
normal leaves. Unknown developmental and/or physiological mechanisms led to this
pattern of insecatus leave phenotype distribution.
In our collection we have not observed the insecatus phenotype in
early flowering pea lines. In order to analyze the influence of a flowering node
on formation of insecatus character the F2 hybrids differed in
lf alleles were studied (Table 1). The lines OK7, OK14 (kindly provided
by Dr. O.E. Kosterin) and WL1325 flower at node 6, counting the first scale leaf
as node 1, and have lf-a, Ins genotype. The lines WL1751 (Ins),
WIR2521 (ins) WIR319 (ins) belong to the late flowering class.
In contrast to homeotic pea leaf development mutants afila and tendril-less(tl)
the mutation of gene ins affects only a few compound pea leaves. We have
observed the insecatus phenotype in several lines and determined the late
flowering the insecatus phenotype appearance 2-3 nodes below the first
flowering node. Following initial formation of insecatus leaves, they may
also occur at the next 3-4 nodes. Thus, insecatus leaves are formed in
the middle part of a plant whereas lower and upper nodes of such plants have
normal leaves. Unknown developmental and/or physiological mechanisms led to this
pattern of insecatus leave phenotype distribution.
In our collection we have not observed the insecatus phenotype in
early flowering pea lines. In order to analyze the influence of a flowering node
on formation of insecatus character the F2 hybrids differed in
lf alleles were studied (Table 1). The lines OK7, OK14 (kindly provided
by Dr. O.E. Kosterin) and WL1325 flower at node 6, counting the first scale leaf
as node 1, and have lf-a, Ins genotype. The lines WL1751 (Ins),
WIR2521 (ins) WIR319 (ins) belong to the late flowering class.
|
Table 1. Comparative analysis of
the first flowering and insecatus nodes in F2 insecatus
plants from different crosses.
|
|
Cross
|
N
|
Flowering
|
First
flowering node ±S.E.
|
First
insecatus node ±S.E.
|
|
WL1325
x WIR2521
|
16
|
Late
|
21.1
±
0.6
|
19.5
±
0.6
|
|
WL1751
x WIR2521
|
35
|
Late
|
19.8
±
0.4
|
16.7
±
0.5
|
|
WIR319
x OK14
|
30
|
Late
|
24.5
±
0.8
|
19.5
±
0.6
|
|
WIR319
x OK7
|
21
|
Late
|
24.9
±
0.9
|
18.7
±
0.6
|
|
WIR319
x OK7
|
03
|
Early
|
6.0
±
0.0
|
15.7
±
2.1
|
Data for all late plants of five crosses presented in Table 1 were used
to obtain a correlation between the first flowering node and the first insecatus
node. The correlation coefficient is r=0.83. These results show a regular trend:
the higher the first flowering node; the higher the first insecatus node.
The interaction of different leaf development mutants is of great
interest because of the control of compound leaf development. We described the tlw
gene influence on exhibition of insecatus phenotype. Homozygous tlw/tlw,
ins/ins plants had instead of overdeveloped central vein a small leaflet
(Fig. 1, middle). In heterozygous Tl/tlw plants the
overdeveloped central vein looked like a flat tendril (Fig. 1, right). Thus, a
form of central vein of insecatus plants is completely defined by gene tl.
The gene ins itself is responsible for extension of central vein beyond
the edge of the blade and more or less symmetric leaflet dissection on both
sides of this vein.
Lamprecht established a single recessive gene inheritance for insecatus
leaves (1). We have analyzed five hybrid combinations differing for ins
alleles. In our experiments all F1 plants had wild type phenotype. F2
plants with any degree of dissection were identified as a recessive
homozygous class. With such an approach the number of plants in different
phenotypic classes is in a good accordance with the theoretically expected 3:1
ratio for the monogenic recessive control of insecatus leaves.
Encouraged by the good 3:1 fit for Ins – ins segregation in F2
we have studied segregation of ins with marker genes. The fact that ins
was in repulsion phase relative to the most of marker genes made the
interpretation of segregations data more complicated because of large standard
errors for the determination of genetic distances.
There was no linkage between ins and any of 29 marker genes listed
here: d, o, i, ar, M, b, gl, v,
gp, te, Fs, Ust, Pl, r, tlw,
bt, le, Np, fas, k, wb, a, lf,
blb(2), oh, st, SCA(3), PSP3, PSP7 (the
two last code proteins separated by electrophoresis in acetic acid). Lamprecht
also failed to locate ins (1). In his experiments the initial pea line
has the gene ins with precise expressivity, while in the F2
progeny the plants appeared with insignificant leaflet dissection. The
continuous variability from strong to insignificant dissection created
complexities at phenotypic classification.
 Our experiments also show there are genes modifying the gene ins
expression. Two lines WIR319 and WL6 each with the "normal" insecatus
expressivity (5-6 incised leaflets per a plant) have been crossed. All F2
offspring had dissected leaflets but the amount of insecatus leaflets
varied very wide from plant to plant. As all F2 plants were recessive
homozygotes ins/ins and were growing in the same conditions of a
greenhouse, it is obvious that a genetic background of a plant influences on an insecatus
gene expression.
Our experiments also show there are genes modifying the gene ins
expression. Two lines WIR319 and WL6 each with the "normal" insecatus
expressivity (5-6 incised leaflets per a plant) have been crossed. All F2
offspring had dissected leaflets but the amount of insecatus leaflets
varied very wide from plant to plant. As all F2 plants were recessive
homozygotes ins/ins and were growing in the same conditions of a
greenhouse, it is obvious that a genetic background of a plant influences on an insecatus
gene expression.
In
some case the mutation ins, like ins2 (4, 5), displays dominance.
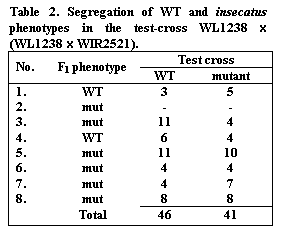
In the cross WL1238 x WIR2521 we have obtained 8 F1
plants: 2 F1 with WT and 6 F1 with mutant insecatus
phenotype. Seven F1 plants were used to generate the test-cross
WL1238 x (WL1238xWIR2521) (Table 2).
Test-cross data reveal good 1:1 segregation. But varying dominance of insecatus
phenotype in the same cross makes genetic mapping of ins gene difficult.
For further genetic analysis, we will need to find ins alleles with
better penetrance and stable expression.
1.
Lamprecht, H. 1959. Agri Hort. Genet. 17: 26-36.
2.
Kosterin, O.E., Rozov, S.M. 1993.
Pisum Genetics 25: 27-31.
3.
Smirnova, O.G., Rozov, S.M., Kosterin, O.E. and Berdnikov, V.A. 1992. Plant Sci.
82: 1-13.
4.
Berdnikov, V.A., Gorel, F.L., Bogdanova, V.S. and Kosterin, O.E. 2000. Pisum
Genetics 32: 9-12.
5.
Berdnikov, V.A. and Gorel, F.L. 2001.
Pisum Genetics 33: 26.
 In contrast to homeotic pea leaf development mutants afila and tendril-less(tl)
the mutation of gene ins affects only a few compound pea leaves. We have
observed the insecatus phenotype in several lines and determined the late
flowering the insecatus phenotype appearance 2-3 nodes below the first
flowering node. Following initial formation of insecatus leaves, they may
also occur at the next 3-4 nodes. Thus, insecatus leaves are formed in
the middle part of a plant whereas lower and upper nodes of such plants have
normal leaves. Unknown developmental and/or physiological mechanisms led to this
pattern of insecatus leave phenotype distribution.
In our collection we have not observed the insecatus phenotype in
early flowering pea lines. In order to analyze the influence of a flowering node
on formation of insecatus character the F2 hybrids differed in
lf alleles were studied (Table 1). The lines OK7, OK14 (kindly provided
by Dr. O.E. Kosterin) and WL1325 flower at node 6, counting the first scale leaf
as node 1, and have lf-a, Ins genotype. The lines WL1751 (Ins),
WIR2521 (ins) WIR319 (ins) belong to the late flowering class.
In contrast to homeotic pea leaf development mutants afila and tendril-less(tl)
the mutation of gene ins affects only a few compound pea leaves. We have
observed the insecatus phenotype in several lines and determined the late
flowering the insecatus phenotype appearance 2-3 nodes below the first
flowering node. Following initial formation of insecatus leaves, they may
also occur at the next 3-4 nodes. Thus, insecatus leaves are formed in
the middle part of a plant whereas lower and upper nodes of such plants have
normal leaves. Unknown developmental and/or physiological mechanisms led to this
pattern of insecatus leave phenotype distribution.
In our collection we have not observed the insecatus phenotype in
early flowering pea lines. In order to analyze the influence of a flowering node
on formation of insecatus character the F2 hybrids differed in
lf alleles were studied (Table 1). The lines OK7, OK14 (kindly provided
by Dr. O.E. Kosterin) and WL1325 flower at node 6, counting the first scale leaf
as node 1, and have lf-a, Ins genotype. The lines WL1751 (Ins),
WIR2521 (ins) WIR319 (ins) belong to the late flowering class.
