Identification of AFLP markers for the powdery mildew resistance gene er2 in pea
| Tiwari, K.R., Penner, G.A. and | Agriculture and Agri-Food Canada, Cereal Research Centre |
| T.D. Warkentin | Agriculture and Agri-Food Canada, Morden Research Centre |
Current address of TDW: Crop Development Centre, University of Saskatchewan, 51 Campus Drive, Saskatoon, SK, Canada, S7N 5A8, email: warkentin@sask.usask.ca.
Introduction
Novel marker technologies such as random amplified polymorphic DNA (RAPD) and microsatellites became available with the introduction of polymerase chain reaction (PCR). A relatively new marker system, amplified fragment length polymorphism (AFLP) (10) has been developed where the reliability of restriction fragment length polymorphism (RFLP) is combined with the power of the PCR technique (10); however, AFLPs are dominant markers. This technique has been successfully used to identify markers for disease resistance genes (5).
Powdery mildew, caused by the obligate parasite Erysiphe pisi Syd. (Syn. E. polygoni DC.), is a serious disease of pea (Pisum sativum L.), reducing both yield and quality. Resistance to this pathogen is controlled by recessive gene(s) er1 and/or er2 (2, 8). RAPD markers closely linked to er1 have been identified (6, 7). Pea line JI 2480 has been previously shown to carry er2 (8). Combining both resistance genes er1 and er2 in a cultivar could increase the durability of resistance. Identification of molecular markers for er2 would facilitate the introgression of this gene in lines carrying er1. Therefore, the objective of this study was to identify markers linked to er2.
Materials and methods
Plant materials
Crosses were made between a resistant line JI 2480 (er2) and a susceptible cultivar Radley in 1994. A fraction of the F
1 seed was grown in a greenhouse to produce the F2. Parents, F1 and the F2 progeny were screened under field conditions at the Agriculture and Agri-Food Canada, Morden Research Centre, Morden, Manitoba in 1995 to determine the disease reaction of individual plants. F2-derived F3 families were grown under field conditions at the Morden Research Centre in 1996. Natural infection by powdery mildew occurred in both years. Disease reaction exhibited by the F3 families of selfed F2 plants was used to determine homozygous susceptible and resistant lines. A total of 42 homozygous resistant and 39 homozygous susceptible lines were used to screen AFLP primers. Genomic DNA was extracted from freeze-dried leaflets or stipules of two to three-week-old seedlings by a cetyltrimethylammonium bromide (CTAB) method and quantified using a Spectronic Genesys 5 (Milton Roy) spectrophotometer.AFLP analysis
The AFLP procedure was performed following the protocol of Vos et al. (10) with minor modifications. From each sample, one microgram (µg) of genomic DNA was double digested with 5 units (u) each of EcoRI (Boehringer Manheim) and MseI (BRL). Then, two picomoles of EcoRI adapters, 20 picomoles of MseI adapters were ligated. The working solution was diluted to 5 nanogram (ng)/microliter (µL) with TE
0.1 (10 mM Tris-HCl, 0.1 mM EDTA (pH 7.5). The first PCR (pre-amplification) was performed with one selective nucleotide (EcoRI +A, MseI + C). Each reaction consisted of 1x Promega Biotech Taq activity buffer, 1.5 mM MgCl2, 1 u Taq DNA polymerase, 800 µM total dNTPs, 30 ng of EcoRI primer and 30 ng of MseI primer with 5 ng of genomic DNA. Pre-amplified solutions were diluted 10 fold in TE0.1 and selective amplification was performed with the EcoRI primer + A+ 2 selective nucleotides and MseI primer + C+ 2 selective nucleotides. Two separate DNA pools were prepared from eight homozygous resistant plants and eight homozygous susceptible plants from different F3 families. Each pool contained an equal amount of pre-amplified DNA among the individuals. All possible primer combinations between 8 EcoRI (AAC, AAG, ACA, ACC, ACG, ACT, AGC, AGG) and 16 MseI (CAA, CAC, CAG, CAT, CCA, CCC, CCG, CCT, CGA, CGC, CGG, CGT, CTA, CTC, CTG, CTT) primers were screened between the pools with a total of 128 primer combinations.Electrophoresis in polyacrylamide denaturing gels and silver staining
Following amplification, reaction products were mixed with an equal volume (20 µL) stop solution (98% formamide, 10 mM EDTA pH 8.0, and bromophenol blue and xylene cyanol). The resulting mixtures were denatured and samples (2.5 µL each) were loaded in a 5% sequencing gel. The gel was pre-run for 45 minutes at 85 W in 1x TBE and electrophoresed at constant power of 85 W for 2.5 h. Following electrophoresis, gels were silver stained following the Promega silver sequence
TM protocol. Images were stored by scanning with image capture software, Photostyler.Results and Discussion
AFLP analysis appears to be promising for genetic studies of pea. A total of 40 to 80 DNA bands per lane were evident in an AFLP denaturing polyacrylamide gel as compared to two to eight bands in a RAPD analysis followed by agarose gel electrophoresis. These data indicate that AFLPs have a clear advantage over RAPDs in terms of number of amplicons amplified per reaction. Out of 128 primer combinations screened between the powdery mildew resistant and powdery mildew susceptible bulks, amplicons of ten primer combinations showed a 0% to 20% recombination with er2 upon screening individual lines which constituted the bulks. The entire population of 81 individual lines was screened with these ten primer combinations; amplicons of four primer combinations were closely linked to er2.
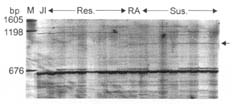
EcoRI primer 5'- GACTGCGTACCAATTC-3' (E) + three selective nucleotides and MseI primer 5'-GATGAGTCCTGAGTAA-3' (M) + three selective nucleotides were used to screen the population from the cross JI 2480/Radley. The primer combination E+ACT (selective nucleotides)/M+CGC amplified a polymorphic fragment of approximately 1000 base pairs (bp) in the susceptible parent Radley (Fig. 1). Out of 81 progeny, the fragment was present in eight of the resistant progeny (8/42) and absent in three of the susceptible progeny (3/39) indicating a linkage distance of 11/81= 14 ± 4 cM. The primer combination E+ACG/M+CCC amplified a polymorphic fragment of approximately 460 bp in the susceptible parent Radley. The fragment was present in seven of the resistant progeny and absent in three of the susceptible progeny (12 ± 4 cM) (figure not shown). Similarly, an amplicon of the primer combination E+AGG/M+CTA amplified a polymorphic amplicon of 241 bp in the susceptible parent Radley. This amplicon was present in two of the resistant progeny and absent in two of the susceptible progeny (5 ± 2 cM) (figure not shown).

Fig. 1. Polymorphic amplicon (~1000 bp) amplified by primer combination 5’-GACTGCGTACCAA TTCACT and 5’-GATGAGTCCTGAGTAACGC. JI=JI 2480, RA=Radley. Res.=Resistant progeny, Sus.=Susceptible progeny. M=Molecular weight marker. The arrow on the right indicates the polymorphic band.
The primer combination E+AGG/M+CTG amplified two polymorphic fragments in cis to er2 in resistant parent JI 2480. One of the fragments was approximately 600 bp (Fig. 2). The fragment was absent in two of the resistant progeny and present in six of the susceptible progeny indicating a linkage distance of 10 ± 3 cM. The second amplicon of this primer combination was 123 bp and the fragment was absent in three of the resistant progeny and present in five of the susceptible progeny (10 ± 3 cM) (figure not shown).
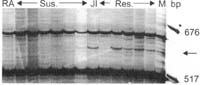
Fig. 2. Polymorphic amplicon (~600 bp) amplified by primer combination 5’-GACTGCGTACCAATTCAGG and 5’-GATGAGTCCTGAGTAACTG. JI=JI 2480, RA=Radley. Res.=Resistant progeny, Sus.=Susceptible progeny. M=Molecular weight marker. The arrow on the right indicates the polymorphic band.
Although er1 was identified over fifty years ago (1) and has been used in plant breeding since then, pathogen isolates were detected which were virulent in lines carrying er1 (4, 9). Partial/incomplete resistance, such as that provided by er2 (2, 8), could be more durable than the complete resistance provided by er1 as has been the case in many cereal diseases including barley powdery mildew (3). Incorporation of both genes (er1 and er2) in a cultivar could increase the durability of resistance of both genes. Marker-assisted selection (MAS) would facilitate this process because: i) selection of lines carrying both er1 and er2 on the basis of visual disease scoring would be difficult because er1 alone provides a high level of resistance (2, 8), ii) the obligate parasitic nature of E. pisi makes it difficult to maintain in culture, and iii) disease occurrence under field conditions is uncertain in many regions. We have identified three AFLP markers linked in trans and two AFLP markers linked in cis to er2. The cis phase primer combination E+AGG/M+CTG will be useful in MAS of heterozygous BC
n F1 individuals for JI 2480 derived resistance by the presence of the polymorphic amplicons. The trans phase primer combinations E+ACT/M+CGC, E+ACG/M+CCC, and E+AGG/M+CTA will be useful in identifying homozygous resistant individuals by the absence of the polymorphic amplicons.Acknowledgements: This project was funded by Western Grains Research Foundation (WGRF) for which we are grateful. We greatly appreciate the co-operation of Mike Ambrose (John Innes Institute) and N.F Weeden (Cornell University). We sincerely appreciate the photographic expertise of Reg Sims. We thank G. Casey, L. Bezte, S. J. Lee, A. Sloan, and B. Gillis for their help in the laboratory and field work.
0
1. Harland, S.C. 1948. Heredity 2:263-269.