Two simultaneously induced lethal mutations provide a system for automatic reproduction of a heterozygote for the Hammarlund translocation
| Berdnikov, V.A., Gorel, F.L. and Kosterin, O.E. |
Institute of Cytology and Genetics Novosibirsk 630090, Russia |
In a paper by Gorel et al. (1) in this issue an approach was described for obtaining mutations linked to the breakpoint of the Hammarlund translocation. We have treated the hybrid Tau2 with EMS and obtained M3 families resulting from heterozygotes for Hammarlund translocation (1). In one of those families we found a plant (A Cri Gp) which had about 33% immature, bubble-like seeds. The testa of these bubble-like seeds looked normal, but inside the testa was considerable liquid, and the embryos were extremely retarded (usually only 2 mm in diameter). From this plant, which was the source of the line VEL, we gathered 17 normal seeds and grew them hydroponically in a greenhouse.
We expected four well-defined phenotypic classes: 1) semisterile heterozygotes (K/N), similar to the parent plant, with the phenotype Cri Gp A and heterozygous for haplotypes 2121 and 1123 of the histone H1 gene cluster His(2-6); 2) fertile homozygotes for translocation (K/K), with the phenotype Cri Gp A and a haplotype 2121 of His(2-6); 3) fertile homozygotes for normal karyotype (N/N) with phenotype a cri gp 1123, and 4) tertiary trisomics N/N+5
2 with phenotype a Cri gp 1123 (1). However, the progeny of the line VEL founder consisted only of two phenotypic classes. Eight plants were weak and slowly developing, possessing pale-green foliage with a network of darker veins. The other nine plants were large with normal foliage, although they flowered and ripened rather late and gave relatively few seeds. From a single “normal” plant, selfing was continued for six generations to generate the line VEL. The data concerning phenotypes of each generation are presented in Table 1. Table 1. Joint segregation for the phenotypes of the gene cos1 and haplotypes of the histone H1 gene cluster His(2-6) in different generations of selfing of a line VEL.| Generations selfing after M3 |
cos1 2121 |
cos1 2121+1123 |
Cos1 2121+1123 |
| G1 | 8 | 0 | 9 |
| G2 | 7 | 0 | 13 |
| G3 | 12 | 1 | 7 |
| G4 | 4 | 0 | 18 |
| G5 | 11 | 1 | 17 |
| G6 | 8 | 0 | 7 |
| Total | 50 | 2 | 71 |
The phenotype of the pale plants resembled that of the mutation class costata, usually designated as lum with numerals. To date, five mutations of this class have been described by different authors (see the following comment by Gene Symbol Coordinator) and three have been mapped. At present the nomenclature of the lum loci and their type lines is uncertain. We therefore are following the recommendation of the Coordinator and calling our mutation cos1 on a provisional basis. All cos1 plants were homozygous for the His(2-6) haplotype 2121 and, despite reduced vigor, possessed fertile pollen. All normal plants with the phenotype Cos1 were determined to be structural heterozygotes K/N, their histone phenotype was 1123+2121, their pollen was semisterile, and some seeds in their pods had a bubble-like appearance. From ripened seeds gathered from heterozygotes in the course of six generations of self-pollination, 123 plants were obtained, among which 52 had phenotype cos1. Only two cos1 plants were heterozygous for histone gene His(2-6), the remaining 50 cos1 plants were homozygous 2121/2121. It should be noted that cos1 plants are so weak that they perish in the field at the stage of 5-6 nodes giving no flowers. All 71 plants with the normal foliage color were structural heterozygotes like their parents. Structural homozygotes with phenotype cri gp a 1123 were absent, tertiary trsomics N/N+5
2 with phenotype Cri gp a 1123 were absent as well. All data suggest that a recessive sporophytic lethal is situated on either chromosome* 2 or 5 of the normal karyotype, the presence of bubble-like seeds may be ascribed to action of this lethal. H1 histone isolation was performed from the embryos of bubble-like seeds, the formula of His(2-6) haplotype proved to be 1123, consistent with the above suggestion. We called this embryonic lethal “vesicula“, ves, (‘a bubble‘ in Latin). The complete absence of trisomics N/N+52 indicated that the lethal ves is not covered by the short interchange chromosome 52 and must be situated in the long arm of either chromosome 2 or 5.One normal Cos1 plant from the sixth generation of VEL was crossed with line SG-802 (normal karyotype A Cri Gp His(2-6)
1121). From the F1 progeny one plant with heterozygous histone H1 phenotype 1221+1123 was selected. The high fertility of this plant implied that it possessed a normal (or at least balanced) karyotype. Phenotypes of 241 F2 progenies are presented in Table 2. A strong deficit of plants with phenotype gp (only one plant instead of expected 60) is evident, suggesting a tight linkage between lethal ves with gp. As the locus Gp is linked to Cri, the lack of plants with phenotype cri can also be explained by linkage with the lethal. The single gp plant must be heterozygote ves/+, arising as a result of cross-over event between Gp and Ves. The fact that the gp plant was homozygous for cri indicates that the locus Ves is situated on chromosome 5 distal to Gp. The data of Table 2 allowed us to calculate the distances between the lethal and the markers cri and gp using the maximum likelihood method (see ref. 2) as follows:lethal-cri: R.f. = 8.45, S.E. = 1.38, joint chi-square = 49.4,
P(0.5) < 0.0001
lethal-gp: R.f. = 0.62, S.E. = 0.36; joint chi-square = 77.6; P(0.5) < 0.0001
| Cri Gp | cri Gp | Cri gp | cri gp | |
| A | 210 | 12 | 0 | 1 |
| a | 18 | 1 | 0 | 0 |
that resulted in the following map segment:
cri ———————————————————gp—ves
8.45 cM 0.62 cMx
The obvious shortage of plants with phenotype a is difficult to attribute to effects of the lethal ves because ves resides on a different chromosome.
It follows from Table 2 that the proportion of cos1 plants in the progeny of structural heterozygotes reaches 40 percent. Among 52 plants only two with this phenotype possessed a heterozygous haplotype of the histone H1 gene cluster His(2-6), the rest being homozygotes 2121/2121. This result indicates that mutation cos1 resides in one of interchange chromosomes 5
2 or 25. We crossed structural heterozygote of line VEL with our original line HT3. This line has a translocation karyotype K/K with the long interchange chromosomes marked by His(2-6) haplotype 2121 and recessive alleles lfa a gp. A plant with phenotype 2121 Lf A Gp Cos1 was chosen from the F1, which must have been the structural homozygote K/K. Phenotypes of 141 progenies are given in Table 3. Table 3. Phenotype segregation in F2 progeny of an F1 plant with a normal karyotype and a phenotype Lf A Gp Cos1 resulting from a cross [VEL heterozygote x HT3 (N/N gp lf)]GeneA |
GeneB |
Phase |
A/B |
A/b |
a/B |
a/b |
Total |
Linkage % |
St.Error % |
Joint Ch.sq. |
P(0.5) |
| cos1 | gp | Rep. | 75 | 41 | 24 | 0 | 140 | Uncertain | Uncertain | 12.00 | <0.001 |
| Cos1 | lf | Rep. | 81 | 36 | 21 | 3 | 141 | 33.90 | 7.35 | 3.32 | <0.1 |
| Gp | a | Coup. | 83 | 16 | 18 | 23 | 140 | 26.15 | 4.45 | 23.01 | <0.0001 |
| a | lf | Coup. | 90 | 5 | 12 | 34 | 141 | 11.86 | 2.93 | 73.00 | <0.0001 |
Calculations were made using the maximum likelihood method with the aid of the programs PLANTS and CROS.
Among 24 plants with phenotype cos1, four plants had white flowers, but none had the phenotype gp. Bearing in mind the repulsion phase of cos1 and recessive markers, this indicates a closer linkage between Cos1 and Gp than between Cos1 and either A or Lf. The data in Table 3 permit us to estimate the recombination rate between loci A and Gp on the long interchange chromosome (in homozygotes for the translocation karyotype K/K) to be 18 percent. The recombination rate between the same markers (separated by T-points) in the karyotype K/M is about 1 per cent, indicating that structural heterozygosity reduces the rate of recombination in the vicinity of this T-point more than ten-fold! The map distance between the loci A and Lf does not differ greatly from that of the normal karyotype. In addition, the deficit of cos1 plants is obvious. Remembering the similar deficit of plants with phenotype a in the F
2 shown in Table 2, one can suppose that all the chromosomes involved in the translocation and derived from the line VEL carry some harmful mutations.The plants with phenotype lf
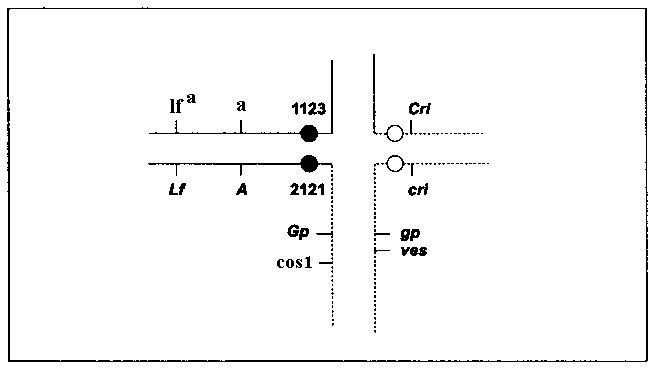
a a cos1 are the products of combining of two recombinant chromosomes resulting from cross-over events between the loci Cos1 and A. If Cos1 lies distal to Gp, than one of cross-over events could occur between Cos1 and Gp. The analysis of the progeny of self-pollination of three a lfa cos1 plants revealed that one plant was heterozygous at the locus Gp, as one of its seven off-springs had the phenotype gp. This finding proves that Cos1 is distal to Gp. It is known that the gene lum-3, sensu Swiecicki (4), with similar effect on foliage (3), is situated distal to Gp on the long arm of chromosome 5. We crossed the line RT-9, carrying lum-3, with a cos1 plant, but the hybrid had a normal color of foliage, demonstrating non-allelism between cos1 and lum3.Thus, the structural heterozygote of the line VEL carries two mutations located not far from T-points of the Hammarlund translocation (Fig. 1). One of them, an embryonic lethal ves, resides in the long arm of chromosome 5 of the normal karyotype slightly distal to Gp. The other mutation, cos1, dramatically reducing the fitness of the sporophyte, lies on the homologous arm of the longer interchange chromosome 2
5 distal to Gp. It is surprising that both harmful mutations were simultaneously induced by EMS in homologous regions of different chromosome sets. As cos1 plants perish in the field, in the case of line VEL we have the balanced system of lethals, providing the automatic reproduction of heterozygosity for the substantial segment of the pea genome.
Fig. 1. A scheme of a cross formed in meiosis by translocated chromosomes and their normal homologs in a line VEL, heterozygous for Hammarlund’s translocation and bearing balanced lethal genes ves and cos1.
Acknowledgement: We express our gratitude to Mike Ambrose for valuable comments and information on nomenclature of the costata-type mutations. This work was partly supported by the Russian State Program, “Russian Fund for Fundamental Research.”
1. Gorel, F.L., Kosterin, O.E. and Berdnikov, V.A. 1999. Pisum Genetics 31:5-8.
2. Kosterin, O.E., Berdnikov, V.A. and Gorel, F.L. 1999. Pisum Genetics 31:33.
3. Rozov, S.M. 1993. Pisum Genetics 25: 43-4.
4. Swiecicki, W.K. 1988. PNL 20: 36-37.