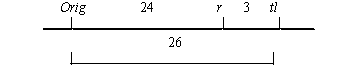
A probable codominant allele of the locus bt with a striking effect on the pod
Kosterin, O.E., Berdnikov, V.A.,
Institute of Cytology
and Genetics
Rozov, S.M. and Gorel
F.L. Novosibirsk
630090, Russia
In 1970, Sobolev and Bugrii (3) reported on a pea with a striking pod shape. In 1961, V.P. Bugrii found an anomalous oblong seed among those obtained from the cultivar Sever (Vulgatum 63) in the field in the Tomsk region. The resulting plant had very long, narrow and curved pods which had inside an extensive spongy layer; the pods usually cracked open before the seeds ripened. The progeny from this plant segregated for the characters in question, their expression being highly variable. Bugriy gave this syndrome the name “Originalnyi” (which means “peculiar, original”).
Thanks to the courtesy of Dr Alexei Kravchenko, we obtained seeds of the “hybrid Originalnyi”. The seeds were of both round and oblong shape, heterogeneous for green and yellow cotyledon colour (the gene i), and lacked anthocyanin pigmentation. We planted 10 seeds which produced plants falling into three phenotypic classes.
i) Three plants had normal pods.
ii) Five plants had long pods curved in a sigmoid manner, with increased distance between the ovules and elongated tips ending with a small hook. In the distal region of some pods, the ventral suture split open during seed development exposing the seeds which finished their growth in the open air. The pods had an extensive spongy layer inside. The first pods on the plants were closer to normal while the later developing pods expressed the abnormalities more strongly. The pods on lateral branches were the most narrow and curved. The narrow pods produced oblong seeds.
iii) Two plants had extraordinarily narrow pods with great interovular distances. The pods were either straight or strongly curved upwards or twisted, with the tip strongly bent ventrally. Almost all pods split open and most of the seeds developed in an exposed state. The spongy layer inside the pod was strongly expressed. Many ovules were sterile or gave abortive seeds; the viable seeds were of a cylindrical shape.
This three-phenotype situation might be explained by the presence of a codominant factor which gives phenotype ii) in a heterozygous state and phenotype iii) when homozygous. Several experiments proved this supposition.
First, we planted the progeny produced by selfing of a single plant with phenotype ii) and obtained 9 plants of phenotype i), 25 plants of phenotype ii), and 5 plants of phenotype iii). Next, we crossed a plant with phenotype iii) with one of our original testerlines “fas bt n”. The F2 progeny from a single fully fertile F1 plant with phenotype ii) consisted of 3 plants of phenotype i), 12 plants of phenotype ii) and 9 plants of phenotype iii) (Fig. 1). If the data for the two segregating progenies are pooled, we obtain the ratio 12:37:14, that is quite close to 1:2:1 (Chi-sq 1.35; P > 0.20). We concluded that the pod syndrome was determined by a codominant genetic factor and gave it a preliminary name Orig.
To attempt linkage analysis, we crossed a plant homozygous for Orig with the testerline WL1238 (JI73) and obtained a single vigorous fully fertile F1 plant which exhibited a phenotype (ii) heterozygous for Orig. 91 F2 plants were grown. They revealed linkage of Orig with markers r and tl of linkage group V (Table 1). The results implied the following linkage relationship:
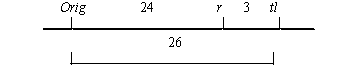
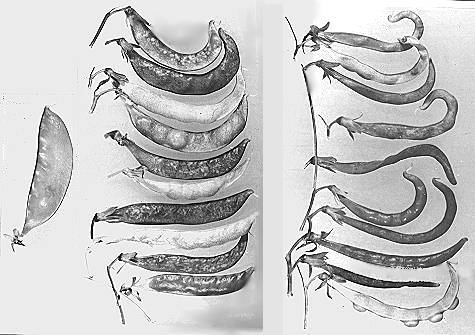
Fig. 1. Pods of F2 plants from the cross between a plant heterozygous for Orig from the initial stock and testerline “fas bt n”. Left, normal; centre, heterozygous for Orig; right, homozygous for Orig.
Table 1. Joint segregation data for the F2 progeny of a single F1 plant from a cross between an Orig plant and testerline WL1238 (r tl).
|
Gene pair |
Number of progeny with phenotype1 |
Joint Chi-sq2 |
RCV2 |
SE |
||||||
|
AB |
Ah |
Ab |
aB |
ah |
ab |
|||||
| gp | tl |
22 |
30 |
24 |
1 |
8 |
6 |
3.3 |
43.0 |
6.3 |
| Orig | tl |
22 |
26 |
12 |
1 |
12 |
18 |
18.1*** |
26.1 |
5.2 |
| r | tl |
23 |
38 |
3 |
0 |
0 |
27 |
78.1*** |
3.1 |
1.9 |
| gp | Orig |
52 |
-- |
24 |
8 |
-- |
7 |
1.3 |
45.1 |
7.4 |
| gp | r |
55 |
-- |
21 |
9 |
-- |
6 |
0.9 |
44.5 |
7.4 |
| Orig | r |
51 |
-- |
9 |
13 |
-- |
18 |
18.2*** |
24.4 |
5.3 |
***P < 0.0001.
1 A, a – first gene;
B, b – second gene; h – heterozygous. A capital letter stands for dominant
alleles of Gp, R, for the factor Orig, and for allele Tl
of a codominant gene tl. In this experiment, we considered Orig
as a dominant gene and pooled together homozygotes and heterozygotes. All
dominant alleles are in coupling phase. 2Here
and in Table 4 calculations were made with the aid of the program “CROS”
by Dr S.M. Rozov. Note: a strong distortion of monogenic segregations resulted
from loss of plants due to attack by Fusarium wilt, which in this
experiment killed many seedlings, mostly of the phenotype R.
In addition, we crossed a plant heterozygous for Orig from the initial stock (with genotype R His1-2 Sca-s Curl) with our original testerline “det r curl” bearing the linkage group V markers r, His1-1, Sca-f, curl. Among 12 fully fertile F1 plants, 6 had a heterozygous phenotype with respect to Orig. These heterozygous plants were pollinated with the same testerline. The results of this testcross are presented in Tables 2 and 3. They suggest the following relationships:
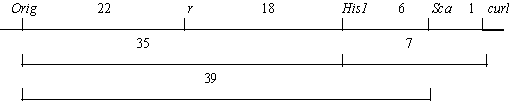
Table 2. Segregation of progeny of the testcross (“Orig” ¥ “det r curl”) ¥ “det r curl”. “Orig” has alleles Orig, R, His1-2, Sca-s, Curl and testerline “det r curl” has alleles orig, r, His1-1, Sca-f, curl*. In the phenotype designations for each locus, symbol + stands for phenotypes corresponding to heterozygotes and symbol – stands for phenotypes corresponding to homozygotes for the testerline alleles. n is the number of plants, their total number being 78.
|
Phenotype for locus |
n |
Phenotype for locus |
n |
||||||||
|
Orig |
r |
His1 |
Sca |
curl |
Orig |
r |
His1 |
Sca |
curl |
||
|
non-recombinant |
non-recombinant |
||||||||||
|
+ |
+ |
+ |
+ |
+ |
23 |
– |
– |
– |
– |
– |
21 |
|
single cross-overs |
single cross-overs |
||||||||||
|
+ |
+ |
+ |
+ |
– |
1 |
– |
– |
– |
– |
+ |
0 |
|
+ |
+ |
+ |
– |
– |
2 |
– |
– |
– |
+ |
+ |
2 |
|
+ |
+ |
– |
– |
– |
3 |
– |
– |
+ |
+ |
+ |
8 |
|
+ |
– |
– |
– |
– |
9 |
– |
+ |
+ |
+ |
+ |
6 |
|
double cross-overs |
double cross-overs |
||||||||||
|
+ |
+ |
– |
+ |
+ |
1 |
– |
+ |
– |
– |
– |
1 |
|
+ |
– |
+ |
+ |
+ |
1 |
||||||
*The testerline “det r curl” was obtained by V.A. Berdnikov and F.L. Gorel by combining mutations in the loci r and det, induced by gamma radiation in initial line SG (1), with a mutation in the locus curl induced by EMS in the same initial line (unpublished). The testerline was then purified by eight generations of selfing.
Table 3. Linkage data from the testcross (“Orig” ¥ “det r curl”) ¥ “det r curl” (see Table 2).
|
Gene pair |
Joint Chi-sq |
Prob |
RCV |
SE |
|
| Orig | r |
25.0 |
< 0.0001 |
21.8 |
4.7 |
| Orig | His1 |
7.3 |
< 0.01 |
34.6 |
5.4 |
| Orig | Sca |
4.1 |
< 0.05 |
38.5 |
5.5 |
| Orig | curl |
0.7 |
< 0.5 |
39.7 |
5.5 |
| r | His1 |
32.5 |
< 0.0001 |
17.9 |
4.3 |
| r | Sca |
25.4 |
< 0.0001 |
21.8 |
4.7 |
| r | curl |
23.0 |
< 0.0001 |
23.1 |
4.8 |
| His1 | Sca |
59.3 |
< 0.0001 |
6.4 |
2.8 |
| His1 | curl |
55.8 |
< 0.0001 |
7.7 |
3.0 |
| Sca | curl |
74.1 |
< 0.0001 |
1.3 |
1.3 |
One can see that Orig maps to the region where bt is located. The bt locus also affects the pod shape; recessive allele bt confers an acute pod apex and allele Bt a blunt apex. The question arises is Orig allelic to bt or is it a separate gene linked to the bt locus? Normal plants from the initial stock had the Bt phenotype. If Orig is an allele of bt, then in the progeny of a cross between genotype Orig/Orig and genotype bt/bt we would never observe a plant with phenotype Bt, i.e. only Orig or bt plants would occur. If Orig and bt are different loci (and hence the original stock was homozygous for the allele Bt), then we would expect the phenotype Bt to result from a cross-over event between Orig and Bt.
We crossed an Orig homozygote (with the genotype Orig R Tl His1-2 Sca-s) from the original stock with testerline RT-6 (bt r tl-w His1-1 Sca-f). The results are presented in Table 4. Linkage relationships (shown below) between Orig and markers of linkage group V are similar to those obtained earlier from the testcross data.
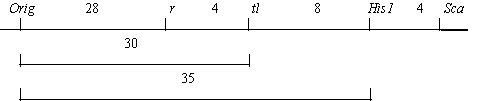
In the F2 of this cross, we observed no plants with the phenotype orig Bt. Of course, the absence of such plants would not confidently prove Orig and bt to be allelic because a possibility remains that they are alleles of very tightly linked loci. But the supposition of allelism seems to be more probable.
Linkage relationships of the linkage group V markers obtained from these two crosses correspond in general to the map obtained by Rozov et al. (2).
Table 4. Joint segregation data for the F2 progeny from the cross of a plant (Orig R Tl His1-2 Sca-s) with the testerline RT-6 (bt r tl-w His1-1 Sca-f).
|
Gene pair |
Number of progeny1 |
Joint Chi-sq |
RCV |
SE |
||||||||||
|
AB |
Ah |
Ab |
hB |
hh |
hb |
aB |
ah |
ab |
||||||
| r | Orig |
16 |
66 |
16 |
-- |
-- |
-- |
2 |
12 |
16 |
16.9** |
27.5 |
4.5 |
|
| tl | Orig |
9 |
19 |
2 |
7 |
44 |
14 |
2 |
15 |
16 |
20.9** |
29.9 |
3.6 |
|
| His1 | Orig |
12 |
16 |
3 |
17 |
45 |
7 |
3 |
17 |
8 |
10.6 |
34.7 |
3.9 |
|
| Orig | Sca |
8 |
6 |
4 |
17 |
44 |
17 |
3 |
15 |
14 |
12.3 |
34.3 |
3.9 |
|
| r | tl |
30 |
64 |
4 |
-- |
-- |
-- |
0 |
1 |
29 |
102.9*** |
3.9 |
1.8 |
|
| r | His1 |
27 |
63 |
8 |
-- |
-- |
-- |
1 |
6 |
23 |
59.0*** |
12.9 |
3.1 |
|
| r | Sca |
27 |
61 |
10 |
-- |
-- |
-- |
1 |
4 |
25 |
61.9*** |
12.8 |
3.1 |
|
| tl | His1 |
25 |
5 |
0 |
2 |
58 |
5 |
1 |
6 |
26 |
153.4*** |
8.2 |
1.8 |
|
| tl | Sca |
25 |
5 |
0 |
2 |
56 |
7 |
1 |
4 |
28 |
154.9*** |
8.2 |
1.8 |
|
| His1 | Sca |
28 |
3 |
0 |
7 |
62 |
0 |
0 |
0 |
28 |
205.4*** |
4.0 |
1.3 |
|
1 A, a – the first gene; B, b – the second gene; h – heterozygous. Alleles from the first parent are denoted with capital letters and those from RT6 with lower case letters. All dominant alleles came from the first parent and are in a coupling phase. **, ***P < 0.001 and 0.0001, respectively. Note: line RT6 was derived by S.M. Rozov and colleagues from lines WL1018 and WL2132 and accessions VIR320 and VIR7036, and purified by six generations of selfing.
In summary, we have observed Orig to behave as a codominant factor located near the end of linkage group V and it is most probably an allele of locus bt. Orig seems not to be accompanied by a structural rearrangement of the chromosome, as crossing-over was not suppressed in the chromosomal region where Orig is located. We do not propose a special gene symbol for Orig and for the present designate it as allele Bt-o. Because Bt-o is located at the extreme end of the known map of linkage group V, it can be useful in genetical work.
Acknowledgements: The authors express their gratitude to Dr A. Kravchenko for providing us with the initial Orig stock, and to L.P. Romkina for technical assistance. The work was partly supported by the Russian State Program “Russian Fund for Fundamental Research”.
______________
1. Gorel, F.L., Berdnikov, V.A. and Temnykh, S.V. 1994. Pisum Genetics 26:16-17.
2. Rozov, S.M., Temnykh, S.V., Gorel, F.L. and Berdnikov, V.A. 1993. Pisum Genetics 25:46-51.
3. Sobolev, N.A. and Bugrii, V.P. 1970. In: Otdalennaya gibridizatsiya rastenii. Zernovye i zernobobovye kultury. [Far Hybridisation in Plants. Corns and Legumes]. Kolos, Moscow, pp. 415-421 [in Russian].
* * * * *