Identification of a
high copy number, retrotransposon-like sequence: results from an unsuccessful PCR search for a pea homologue of the p53 tumour suppressor
gene
Sarup, V.B. and Dept. of
Biological Sciences & Plant Molecular
Stafstrom, J.P. Northern
Pea axillary buds can be interconverted repeatedly between the growing and dormant states of development (19). The growing state, which is induced by decapitating the terminal bud, is characterized by cell proliferation and the accumulation of mRNAs commonly found in growing organs (e.g., ribosomal proteins, histones, MAP kinase, cdc2 kinase and cyclin B; ref. 2). In contrast, the dormant state is closely correlated with cell quiescence. Dormant buds are metabolically active and synthesize many proteins not found in growing buds (18). The identities and functions of dormancy proteins are poorly understood. We reasoned that bud dormancy might result from active inhibition of cell proliferation.
In mammals, the products of tumuor suppressor genes inhibit cell proliferation (14). Mutations in the human p53 tumour suppressor gene are associated with many types of cancer, suggesting that the wild type p53 protein may play a broad, perhaps universal, role in eukaryotic cell cycle regulation (8, 15). Polymerase chain reaction (PCR) technology has been used to clone homologous genes based on conserved amino acid sequences (9). For example, plant cyclins were first cloned using degenerate PCR primers based on conserved "cyclin boxes" (7). Vertebrate p53 proteins contain five highly conserved amino acid domains (6). Our basic strategy for isolating a p53 homologue included: 1) PCR amplification of various DNA templates using oligonucleotide primers based on four of these conserved domains; and 2) analysis of the PCR products by DNA sequencing and by Southern blotting using the human p53 gene as a probe. Using these approaches, we found no evidence for a p53 gene in pea. Surprisingly, a large number of DNA fragments were amplified when three of these four primers were used singly. Several DNA fragments were sequenced and used as probes on genomic Southern blots. One PCR fragment, called psFII-5, is similar to a copia-like retrotransposon and appears to be a highly repetitive genomic element.
Methods
PCR was performed using Taq DNA polymerase (Perkin Elmer/Cetus) and standard concentrations of other reagents (0.2 mM of each dNTP; 2 mM MgCl2; about 1 mg or less template DNA per 100 mL reaction; and 0.1 mM of each primer). Template DNAs tested were pea genomic DNA, cDNA synthesized from polyadenylated RNA, and inserts isolated from two axillary bud cDNA libraries (prepared from growing and dormant buds). Each of 35 cycles consisted of three steps: denaturation (1 min, 94°), annealing (2 min, 40 to 55° for various experiments) and elongation (3 min, 72°). Oligonucleotide primers based on conserved amino acid domains of vertebrate p53 proteins were synthesized; FII and FIII (forward primers) and RIV and RV (reverse primers) were based on domains II, III, IV and V, respectively (ref. 8; Fig. 1). All primers were degenerate for the encoded amino acids. The nucleotide and corresponding amino acid sequences of these primers are listed below (added restriction sites are underlined; nucleotides and amino acids in brackets are alternatives at a given position; N is any nucleotide; and reverse primers were based on the noncoding strand).
FII PheCysGlnLeuAlaLysThrCysPro:
5'-(SalI)-CGTCGACTT[CT]TG[CT]CA[AG][CT]TNGCNAA[AG]ACNTG[CT]CC
FIII ValVal[ArgLys][ArgLys]CysProHisHis:
5'-(BamHI)-CCGGATCCGTNGTN[AC][AG]N[AC][AG]NTG[CT]CCNCA[CT|C
RIV TyrMetCysAsnSerSerCysMetGlyGIyMetAsn:
5'-(SpeI)-
AACTAGT[GA]TTCATNCCNCCCAT[GA]CAN[GC][TA]N[GC][TA][GA]TT[GA]CACAT
[GA]TA
RV ValCysAlaCysProGly:
5'-(EcoRI)-GGAATTCCNGGACANGCACANAC
DNA fragments amplified by PCR were digested with appropriate restriction enzymes and cloned into the pSPORTl plasmid (BRL). In some cases, specific fragments were isolated from ethidium bromide stained agarose gels; in other cases, entire PCR reactions were cloned in order to obtain a random sample of the amplified fragments. Clones were sequenced by the dideoxy method (Sequenase kit, US Biochemical). DNA sequences were compared to the GenBank database using the BLAST search algorithm (1). For Southern blotting of PCR fragments, DNA was separated on a 1.5% agarose gel and transferred to a nylon membrane. Blots were hybridized with a 32P-labeled random-primed probe corresponding to the 1800 bp human p53 cDNA. Blots were washed at moderately low stringency (1X SSC, 1% SDS at 42°C; ref. 17). For genomic Southern blotting, approximately 30 mg of pea genomic DNA was digested with EcoRI or HindIII, separated on a 0.8% agarose gel and transferred to a nylon membrane. Blots hybridized with human p53 probe were washed as described above; blots hybridized with FII-5 or FII-1 probes were washed at moderately high stringency (0.2X SSC, 1% SDS at 58°C).
Results
PCR conditions were optimized
for pairs of forward and reverse primers using the human p53
cDNA clone as a template (data not shown). Pea genomic DNA was amplified using
pairs of forward and reverse primers;
single primers also were used, with the expectation that they would represent a negative control. An agarose gel
of PCR products demonstrates two unexpected
results (Fig. 2). First, even though the primer annealing conditions were moderately stringent (55°C), a large number of
bands was present in nearly all samples. Second, single primers also
amplified large numbers of bands. By examining the original gels, it appears that amplification patterns from a pair
of primers may be merely the sum of amplification
products from the single primers.
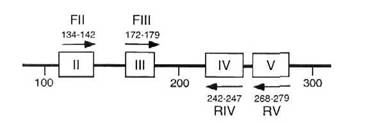
Fig. 1. Schematic representation of a portion of the human p53 protein. Oligonucleotide primers based on amino acid domains II, III, IV and V were synthesized and used for PCR analysis. Arabic numbers indicate amino acid positions.
PCR also was used to amplify DNA inserts from two cDNA libraries and cDNA that was synthesized by reverse transcription of poly-adenylated RNA. Again, large numbers of fragments were amplified, although the dominant bands were not necessarily identical to those amplified from genomic DNA (data not shown). PCR products from all three types of templates were transferred to nylon membranes and hybridized at low stringency with a human p53 probe. Only the positive control (PCR-amplified human p53 cDNA) was recognized strongly by the probe. A few pea bands hybridized very weakly with this probe; we were not successful in subcloning these DNA fragments. Nested PCR also was tried. For this technique, template DNA was amplified with the flanking primers (FII and RV) and the products then were amplified with the internal primers (FIII and RIV). In sum, we were unable to detect any evidence for p53-related sequences in any of these experiments.
PCR amplification using primers FII and RIV was the first experiment performed. Based on the coding region of the human gene, a product of about 340 base pairs would have been expected; however, the size would be larger if introns were present (the human gene contains two introns in this region; ref. 11). Gels contained a strong band at about 300 bases (Fig. 2, arrow). This DNA band was isolated, cloned and several colonies picked for analysis. The sequences of two clones, psFII-1 and psFII-5, were distinct from each other and neither was related to p53 (Figs. 3A and 3B). Both ends of the two clones were similar to the FII primer (in inverted orientation). There are no sequences in the GenBank database that are closely related to clone FII-1. Clone FII-5, however, is quite similar to the yeast TY-3 retrotransposon (135/267 = 51%), the "Bam repeat element" of Vicia aba (184/267 = 69%), and a repetitive sequence element from tobacco (85/131 = 65%). Pea genomic Southern blots were hybridized with psFII-1 and psFII-5 probes (Fig. 4). Several strong bands were detected with the psFII-5 probe after a very short exposure, suggesting that these sequences are highly abundant. Bands hybridizing to the psFII-1 probe could be detected after a longer exposure, but still considerably less time than would be required to detect a single copy gene. Neither sequence could be detected on Southern blots of maize genomic DNA (data not shown).
DNA that was amplified with a single primer (FII, FIII or RV; Fig, 2) was cloned randomly, that is, the entire PCR reaction was digested and ligated into pSPORTl. Two FIII clones, three RV clones and two additional FII clones were analyzed. Each of these clones, which ranged in size from 0.2 to about 1.0 kb, contained the expected "inverted repeat" sequence at both of its ends. The repeats were not identical to each other (see Fig. 4), but when translated, they would correspond to the same degenerate primer. Partial sequencing of the internal portions of these clones did not suggest homology with any previously studied DNA sequences. Southern blots demonstrated that each of these DNA sequences is present in the pea genome but absent from the maize genome (data not shown).
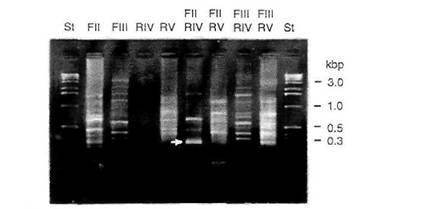
Fig. 2. Agarose gel analysis of PCR amplification products. Pea genomic
DNA was amplified by PCR using single primers or
pairs of forward and reverse primers. St, size standards.
A. Sequence comparisons with PCR clone FII-5
PheCysGlnLeuAlaLysTh rCysPro
FII-5 1 TTCTGTCAATTAGCTAAAAC
GTGCCCGTGCAGAAGAAGGA TGGTAAAGTCAGGATGTGTG 60
TY3-1 2425 CG.CT.GC.GCTC.CCTGTA
..C.T...C.C......A.. C....CCT..C.AC.C..C. 2484
Vicia
534 CG.A..GGG..GC.A.CGTG ..T..G....C......A.C .........TC.T....... 475
N.tab.
123 AGTG..TGGCC.ATGTGGTA ..A..A..C.CA........
...A...A.....G....C. 69
FII-5 61
TCGATTACCGAGATCTAAAC AGAGCTAGTCCGAAAGATGA TTTCCCATTACCTCACATTG 120
TY3-1 2485
..........CACC..G... .A.....CCATCTCC..CCC A...........CAGA..C. 2544
Vicia 474
.A..C..TA.G.....G..T ..G..A.....C..G..... ......TC.G.....T..C. 415
N.tab.
68 ....C.....CA.C..C... .A...A.....C..G..CA. ......G.....CA.....C 9
FII-5 121
ATGTATTGGTTGATAACACG GCTCAATCCTCGGTTTTCTC TTTCATGGATGGATTTTCTG 180
TY3-1
2545 .CAACC.AT.GAGCCGT.TT .GAA.TG..CA.ATA..TA. CACGC.A...TTGCA.AG..
2604
Vicia
414 ....TC.A.....C.....C .TA..GCATAA...C..... C..T........T....T.. 355
N.tab.
8 ..A.CC..A.C 1
FII-5 181
GCTATAATCAGATCAAGATG GCGCCAGAAGACATGGAAAA GACGACATTCATTACACCTT 240
TY3-1
2605 .T..CC.C......CC.... .AA..CA.....CGCT.C.. A..CG.C..TG.C.....A.
2664
Vicia 354
.T...........T...... T.T..C...........G.. ...A..T..TT......AG. 295
ProCysThrLysA1aLeuG1 nCysPhe-N
CCCTGTCCAGAATCGTTCGA
CCGTTTT-5'
FII-5 241 GGGACAGGTCTTAGCAAGCT GGCAAAA
267
TY3-1 2665 CC.GT.A..A.G.AT.TA.C
.T..TGC
2691
Vicia
294 ...GT.CC.T..GCT.CAAA
.T..TGC
268
B. Sequence of PCR clone FII-1
PheCysGlnLeuAlaLysTh rCysPro
FII-l 1 TTTTGTCAACTGGCGAAAAC GTGCCCAGAAACACAATCTC
TATATTATATCAATAAGGAG 60
FII-1
61 GTGTTAGAGGGCCCTCCAAG GTAAGGGTCTACAAGTGGAG
GGACATCATCATGTGCACGG 120
FII-1 121
GAGACGTGCCCCTAGGAAGA TTGCCATTGTGACATGAGGC TCCTAGAAGTCGATGTTTCC 180
FII-1 181
CCGTCAAAAGGCCTAAGGAC GGTTCCCCGCCTAACTTTGG CCTTCTCATTCGGAAGTCCA 240
ProC ysThrLysAlaLeuGlnCys
Phe-N
GCCT
GTTCAGAATCGATCAACTGT TTT-5'
FII-1 241
GCCCCCTCACGTCGAACGGA CAAGTCTTAGCTAGTTGACA AAA 283
Fig. 3. DNA sequence of clones FII-5 and FII-1. A. Comparison of FII-5 with related sequences (dots indicate identity; primer sequences are underlined). TY3-1, Saccharomyces cereviseae TY3-1 retrotransposon (ref. 5; GenBank #M34549); Vicia, Bam repeat element from Vicia faba (ref. 10; GenBank #M16858); N. tab., repeat element from Nicotiana tabacum (unpublished; GenBank #X59606). B. DNA sequence of FII-1. The GenBank database contains no closely related sequences. GenBank accession numbers for clones FII-1 and FII-5 are U25036 and U25035, respectively.
Discussion
The approach used here to look for a pea p53 homologue included PCR amplification using degenerate primers based on conserved domains from vertebrate p53 proteins, detection of related sequences by Southern blotting using a human p53 probe and sequencing the PCR products. Many variations on these procedures were attempted, but none provided any positive evidence for a plant p53 gene. A limited number of related experiments were performed using genomic DNA from maize and Arabidopsis, also without success.
The experiments described here were begun in 1991. At that time, p53 clones had been obtained only from several vertebrate species (8). As of late 1995, there are no reports of p53 genes from well studied animal systems such as Drosophila or Caenorhabditis in the literature or in the DNA databases. Extensive efforts to isolate p53 from budding and fission yeasts also have been unsuccesful, although both species contain proteins that can interact in vivo with human p53 (16, 21). In humans and other vertebrates, the p53 protein appears to be necessary for assuring that DNA is completely replicated and repaired before cell division can occur (6). Budding yeast accomplishes this essential cellular activity using genes/proteins unrelated to p53 (12) and the same may be true of plants.
Results obtained using commercially available p53 antibodies and cDNA probes suggested that p53 protein and mRNA occur in maize embryos (4). Since p53 appears to be found only in vertebrates, these results must be regarded cautiously. Furthermore, the apparent molecular mass of the protein recognized by the antibody was 72 kd (rather than 53 kd) and the sequence that hybridized to the nucleic acid probe has not been cloned.
Three of our four primers, when
used singly, amplified large numbers of DNA fragments (Fig. 2). This result suggested that the ends of the fragments contained
similar sequences in inverted
orientation, which was confirmed for all nine clones that were sequenced. It
remains to be determined whether the corresponding regions of genomic
DNA contain such repeats or whether die
presence of one or both sites in our clones is due to a PCR artifact. Genomic Southern
blots showed that the psFII-1 and psFII-5 clones correspond to moderately or
highly repeated elements (Fig. 4). The fact
that psFII-5 is related to copia-like retrotransposons suggests a mechanism by which it might have been
duplicated. A genomic clone corresponding to psFII-5 might include a complete retrotransposon. Analysis of RFLPs
from various pea accessions and related species might provide evidence
for historical movement of this element
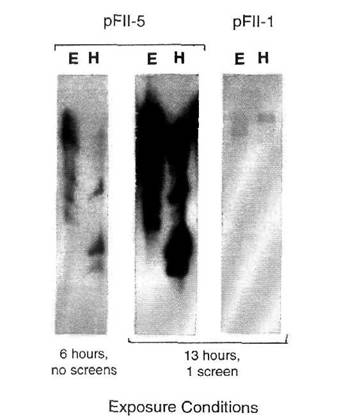
Fig. 4. Southern blot analysis of
pea genomic DNA hybridized with a FII-1 or FII-5 probe. DNA was digested either with EcoRI (E) or HindIII
(H). Exposure conditions are indicated.
within the genome. Copia-like elements are thought to be ubiquitous in plants (20) and have been identified previously in pea (13). The other clones may represent other families of repetitive DNA (3).
Acknowledgments. We thank Dr. Bert Vogelstein,
1. Altschul,
S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. 1990. J. Mol.
Biol. 215:403-410.
2. Devitt, M.L. and Stafstrom, I.P. 1995. Plant Mol. Biol. 29:255-265.
3. Ellis, T.H.N. 1993. In Peas -
Genetics, Molecular Biology and Biotechnology, R.
Casey and D.R. Davies, eds. CAB International, Wallingford, UK; pp 13-47.
4. Georgieva, E.I., Lopez-Rodas, G. and Loidl,
P. 1994. Planta 192:125-129.
5. Hansen, L.J. and Sandmeyer, S.B. 1990. J.
Virol. 64:2599-2607.
6. Harris, C.C. 1993. Science 262:1980-1981.
7. Hirt, H., Mink, M., Pfosser, M., Bogre, L.,
Gyorgyey, J., Jonak, C., Gartner, A.,
Dudits, D. and Heberle-Bors, E. 1992. Plant Cell 4:1531-1538.
8. Hollstein, M., Sidransky, D., Vogelstein, B.
and Harris, C.C. 1991. Science 253:49-
53.
9. Innis, M.A., Gelfand, D.H., Sninsky, J.J. and
White, T.J. 1990. PCR protocols.
Academic Press,
10. Kato, A.,
Iida, Y., Yakura, K. and Tanifuji, S. 1985. Plant Mol. Biol. 5:41-53.
11. Lamb, P.
and Crawford, L. 1986. Mol. Cell Biol. 6:1379-1385.
12. Lydall, D.
and Weinert, T. 1995. Science 270:1488-1491.
13. Lee, D.,
Ellis, T.H.N., Turner, L., Hellens, R.P. and Cleary, W.G. 1990. Plant Mol.
Biol. 15:707-722.
14. Levine,
A.J. 1993. Annu. Rev. Biochem. 62:623-651.
15. Levine,
A.J., Momand, J. and
16. Nigro,
J.M., Sikorski, R., Reed, S.I. and Vogelstein, B. 1992. Mol. Cell. Biol.
12:1357-1365.
17. Sambrook,
J., Fritsch, E.F. and Maniatis, T. 1989. Molecular Cloning, 2nd ed., CSH
Press.
18. Stafstrom,
J.P. and
19. Stafstrom,
J.P. and
20. Voytas,
D.F. 1992. Genetica 86:13-20.
21. Wagner, P., Grimaldi, M. and Jenkins, J.R. 1993. Eur. J. Biochem. 217:731-736.